Functional Genomics and Bioinformatics Core
The Functional Genomics and Bioinformatics Core offers a wide range of Next Generation Sequencing (NGS) technologies and associated bioinformatic analyses to members of the BNORC and Boston area community. This “soup to nuts” approach enables users to go from raw samples to publication-quality figures in one place. Our services can be broadly divided into 3 categories:
Consultation
One of the most important services that the Core provides is pre-experimental consultation. Many investigators know that they want to take advantage of these powerful new tools, but they don’t have the expertise to frame their questions in such a way that matches available techniques. This is a key point: many NGS services are available as send-out services or even as kits that can be purchased directly from vendors and performed in the users’ own laboratories. In our experience, however, the average biomedical scientist is intimidated by the complexity of these techniques. Our Core Directors, technicians, and bioinformaticians have detailed knowledge and hard-won, real-world experience of the pros and cons of available approaches, and the specific challenges of applying these techniques in tissues and cell types relevant to BNORC users.
During consultations, we seek to define and frame experimental questions, then suggest an appropriate experimental approach, taking into account the time, budgetary, or personnel constraints of individual users. The Co-Directors have expertise in the application of each of the core methodologies offered to scientific questions of metabolic relevance and can help users avoid many of the pitfalls that can be encountered, before the commitment of significant time, energy, or money. Of note, in approximately 1/3 of our consultations, we guide users to the conclusion that they do not, in fact, need our services.
Data Generation
In the wet lab, samples are provided by the user in any of a variety of forms, usually frozen tissue, isolated cells, purified RNA or chromatin. Our technical staff then prepare the samples as required using standard operating procedures (SOPs) developed in the Core, and generate libraries. Proper QC is performed at each step along the way. We offer:
- Sample quality assessment of DNA and RNA using the BioAnalyzer and Qubit assays
- Library preparation, sequencing, and analysis of transcriptomes (RNA-seq, including low-input), cistromes (ChIP-seq), chromatin accessibility (ATAC-seq)
- Single Cell/Nucleus isolation, droplet generation and library preparation for RNA-seq, ATAC-seq
- Standalone RNA preparation, RNA and DNA Bioanalyzer analyses, chromatin shearing/shear checks
- Standalone sequencing on NextSeq500
Data Analysis
Each distinct project is assigned to an individual bioinformatician for consistency across that project. For data generated in-house, all data is processed through in-house pipelines that adapt widely-used analysis software (e.g., edgeR, Seurat, CellRanger, CellBender) to provide best practices, high quality, interfaceable data back to the user, including standard PCA, mean-difference, and volcano plots, pathway analysis, and formatted data that can be easily loaded onto platforms such as the Broad Single Cell Portal or Morpheus matrix visualization and analysis. We also provide customized analysis of NGS data, specializing in integration between genetic, transcriptional, and epigenetic datasets.
Equipment
Agilent 2100 Bioanalyzer: The Agilent Bioanalyzer is an established automated electrophoresis tool for the sample quality control of biomolecules. It provides highly precise analytical evaluation of RNA and DNA samples for objective assessment of sizing, quantitation, integrity and purity from RNA input, cDNA synthesis, as well as sequencing library quality and sizing using minimal sample volume.
 Formulatrix MANTIS Liquid Handler: The MANTIS automated liquid handler increases the reproducibility, throughput and efficiency of our RNA-seq workflows. It uses a diaphragm-based chip design and is thus liquid class compatible, dispensing aqueous and organic solutions of varying viscosities down to 100nL at CV < 3%. Efficiency is enhanced via reaction miniaturization as well as reduced dead volumes and tip consumption.
Formulatrix MANTIS Liquid Handler: The MANTIS automated liquid handler increases the reproducibility, throughput and efficiency of our RNA-seq workflows. It uses a diaphragm-based chip design and is thus liquid class compatible, dispensing aqueous and organic solutions of varying viscosities down to 100nL at CV < 3%. Efficiency is enhanced via reaction miniaturization as well as reduced dead volumes and tip consumption.
 10x Genomics Chromium Controller device: The Chromium Controller uses microfluidics to perform single cell partitioning and barcoding in minutes. It is highly efficient (thus reducing RA time), robust for both single cell and single nucleus samples), with increased replicability (easier interface provides reduced variability between experiments), wide applicability across tissue types (has successfully produced data across a spectrum of tissues including nervous system, adipose, pancreas, liver, endothelial cells, muscle), and flexible (stand-alone single-nucleus ATAC-seq as well as simultaneous single cell multi-ome ATAC + Gene Expression kits are now available), with excellent company support.
10x Genomics Chromium Controller device: The Chromium Controller uses microfluidics to perform single cell partitioning and barcoding in minutes. It is highly efficient (thus reducing RA time), robust for both single cell and single nucleus samples), with increased replicability (easier interface provides reduced variability between experiments), wide applicability across tissue types (has successfully produced data across a spectrum of tissues including nervous system, adipose, pancreas, liver, endothelial cells, muscle), and flexible (stand-alone single-nucleus ATAC-seq as well as simultaneous single cell multi-ome ATAC + Gene Expression kits are now available), with excellent company support.
 Miltenyi Biotec GentleMacs Dissociator: The gentleMACS dissociator is a benchtop instrument for semi-automated dissociation of tissues into single-cell suspensions. The gentleMACS Tubes used with the instrument enable the time-saving and reproducible homogenization of tissues in a closed system, and have improved reproducibility our single cell/single nucleus experiments. Specific programs have been developed for various tissues and standardized tissue dissociation and homogenization procedures, enabling reliable and reproducible results.
Miltenyi Biotec GentleMacs Dissociator: The gentleMACS dissociator is a benchtop instrument for semi-automated dissociation of tissues into single-cell suspensions. The gentleMACS Tubes used with the instrument enable the time-saving and reproducible homogenization of tissues in a closed system, and have improved reproducibility our single cell/single nucleus experiments. Specific programs have been developed for various tissues and standardized tissue dissociation and homogenization procedures, enabling reliable and reproducible results.
 Illumina NextSeq 500 high-throughput sequencer. The NextSeq 500 is a high-throughput benchtop sequencer capable of whole-genome sequencing, exome sequencing, RNA-seq, single-cell and single-nuclei RNA-seq, methylation sequencing, and many others. Users can choose between high output or mid output flow cell configurations. At high output, up to 800 million paired-end reads can be generated (at 150 bp read length) to produce up to 120 Gb of data in 29 hours. The Illumina sequencing systems utilize a well-established sequencing by synthesis (SBS) method in which fluorescently labeled nucleotide bases are detected as they are incorporated into DNA template strands. All four reversible terminator-bound dNTPs are present in each sequencing cycle; this feature minimizes bias of incorporation and greatly reduces raw error rates.
Illumina NextSeq 500 high-throughput sequencer. The NextSeq 500 is a high-throughput benchtop sequencer capable of whole-genome sequencing, exome sequencing, RNA-seq, single-cell and single-nuclei RNA-seq, methylation sequencing, and many others. Users can choose between high output or mid output flow cell configurations. At high output, up to 800 million paired-end reads can be generated (at 150 bp read length) to produce up to 120 Gb of data in 29 hours. The Illumina sequencing systems utilize a well-established sequencing by synthesis (SBS) method in which fluorescently labeled nucleotide bases are detected as they are incorporated into DNA template strands. All four reversible terminator-bound dNTPs are present in each sequencing cycle; this feature minimizes bias of incorporation and greatly reduces raw error rates.
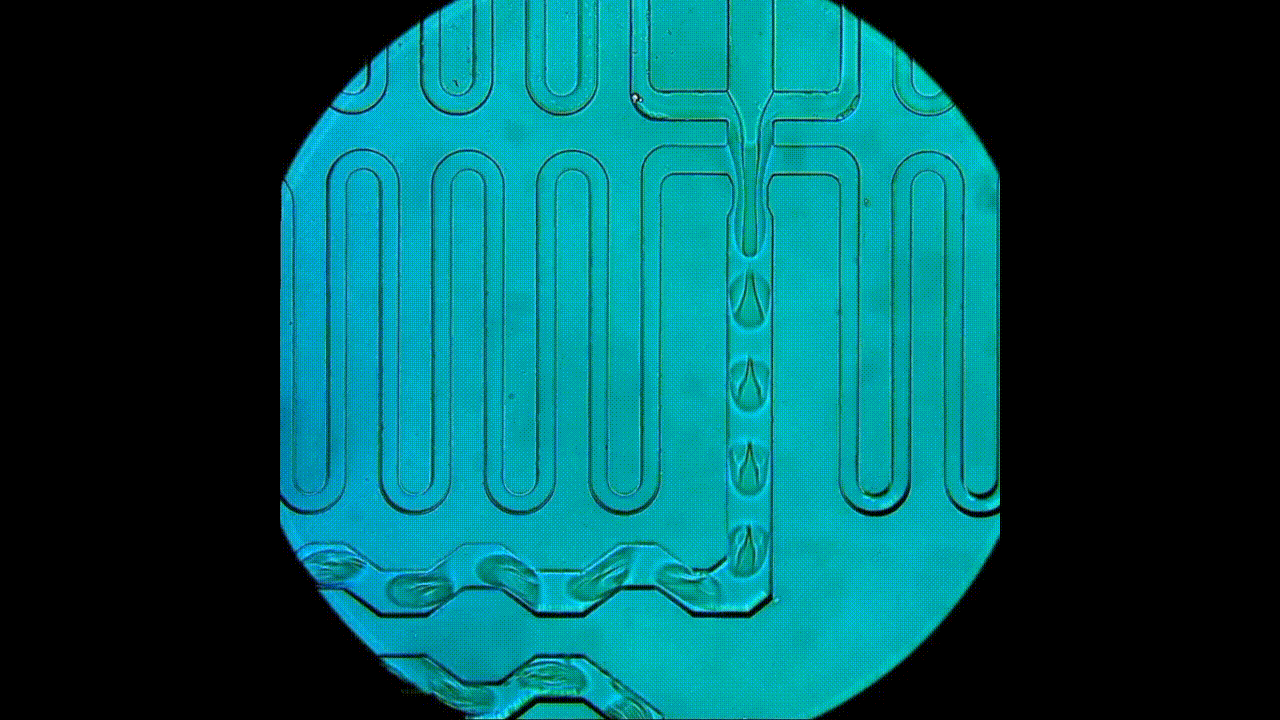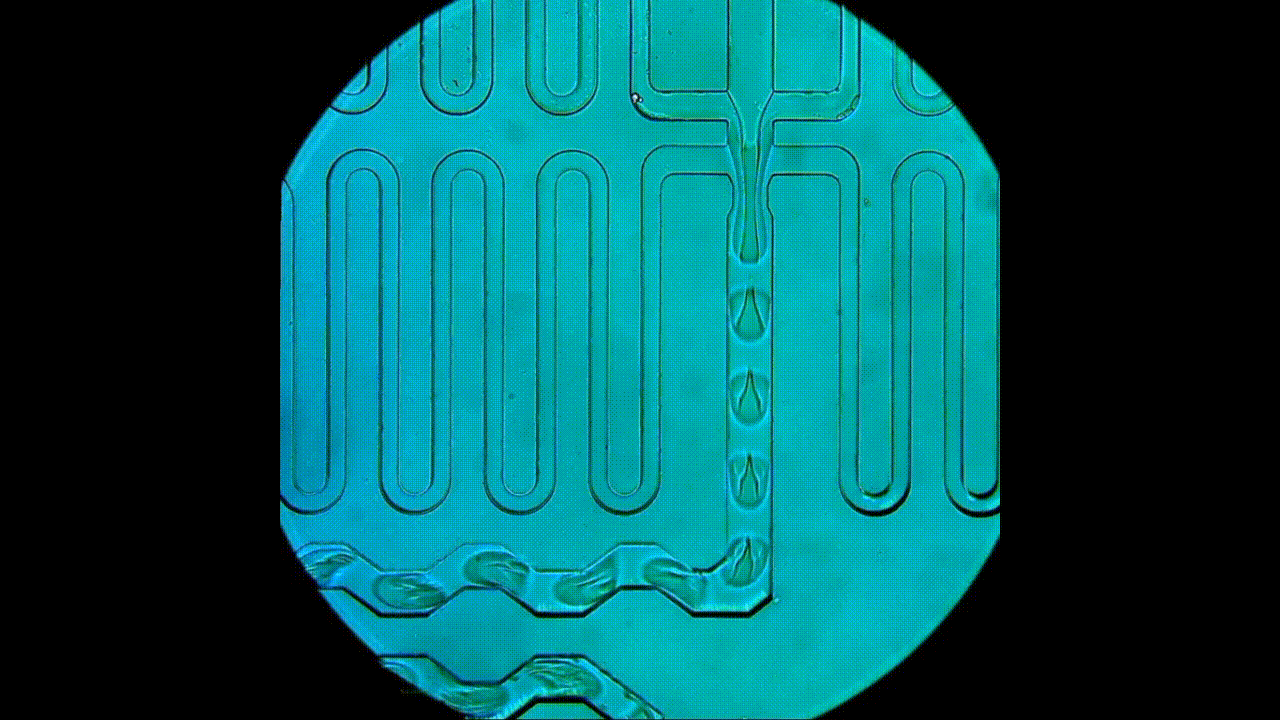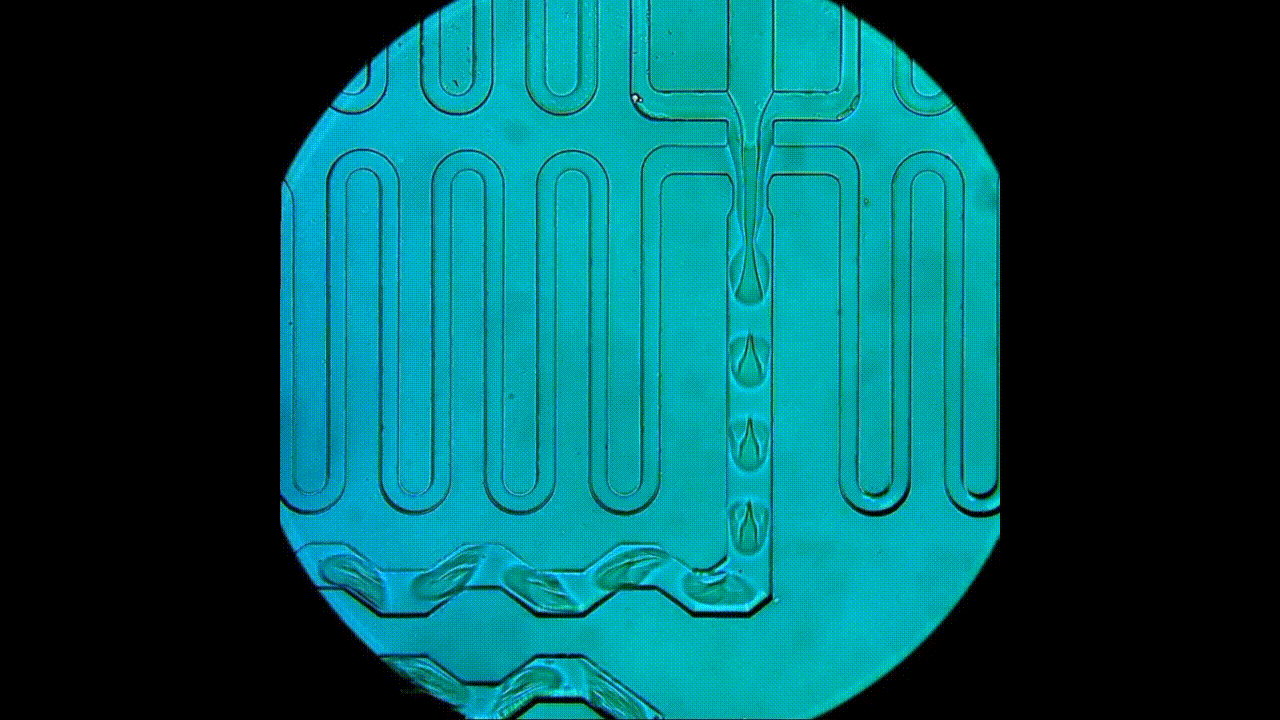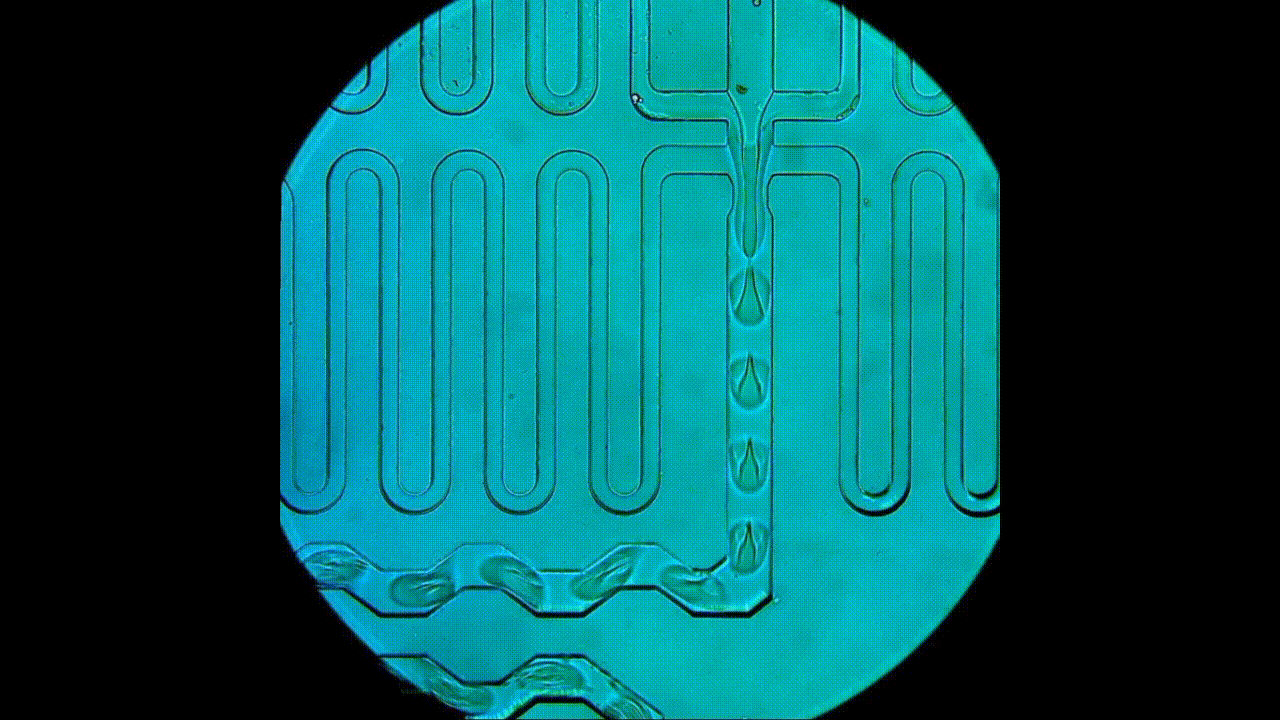 Drop-seq rig for single-cell and single-nucleus transcriptomics. An open-source platform for single-cell/nuclei RNA-seq, this approach allows for smaller input experiments compared to the 10x. The RNA capture beads, microfluidic device, and reagents used for cDNA synthesis are all sourced separately, which provides users with greater flexibility in experimental design and generally lower costs per cell. This apparatus is also used for DroNc-seq (i.e., single nucleus) experiments.
Drop-seq rig for single-cell and single-nucleus transcriptomics. An open-source platform for single-cell/nuclei RNA-seq, this approach allows for smaller input experiments compared to the 10x. The RNA capture beads, microfluidic device, and reagents used for cDNA synthesis are all sourced separately, which provides users with greater flexibility in experimental design and generally lower costs per cell. This apparatus is also used for DroNc-seq (i.e., single nucleus) experiments.
Invitrogen Qubit 3 Fluorometer. Automated system for quantification of DNA and RNA samples. The fluorescent dyes used in the Qubit assays emit signals only when bound to specific target molecules, which minimizes the effects of contaminants, including degraded DNA or RNA. This assay is more sensitive and specific than UV spectrophotometry.
Extensive Q-PCR capabilities (1 Applied Biosystems QuantStudio 5 Real-Time PCR System, and 1 QuantStudio 6 Flex Real-Time PCR System).
Costs
| Project consultation | BNORC | Non-BNORC | Non-Academic |
| Initial project set up | $100 | $150 | $200 |
| Project consultation (hourly) | $150 | $200 | $300 |
| Sequencing Per Flowcell | BNORC | Non-BNORC | Non-Academic |
| NextSeq High 75 cycle | $1,987 | $2,650 | $3,974 |
| NextSeq High 150 cycle | $3,893 | $5,175 | $7,786 |
| NextSeq High 300 cycle | $5,753 | $7,600 | $11,507 |
| BNORC | Non-BNORC | Non-Academic | |
| Full Service Library Preparation - includes QC | per sample | per sample | per sample |
| Crosslinking | $15 | $20 | $30 |
| Shear Check | $150 | $200 | $300 |
| Shearing | $52 | $70 | $104 |
| ChIP setup | $37 | $50 | $74 |
| ChIP antibody | $7 | $10 | $14 |
| DNA, cDNA or ChIP libraries | $236 | $317 | $472 |
| RNA preparation | $25 | $33 | $50 |
| mRNA libraries | $252 | $337 | $504 |
| Poly-A selection | $40 | $53 | $80 |
| Ribo-reduction | $70 | $93 | $140 |
| ATAC-seq | $200 | $267 | $400 |
| If submitting tissue requiring core to setup and perform FACS, charge for FACS will be $150/hr of reserved FACS time | |||
| Single cell/nucleus services | BNORC | Non-BNORC | Non-Academic |
| Drop-seq/Dronc-seq | |||
| Droplet generation: per 2-2.5ml | $600 | $800 | $1,200 |
| Drop-seq Library construction: per ~750 ul run | $600 | $750 | $1,200 |
| 10x single cell RNA-seq | |||
| BNORC* | |||
| Single nucleus suspension prep (per sample) | 1 sample | 2-4 samples | 5-8 samples |
| $360 | $320 | $300 | |
| 10x encapsulation and library construction (per sample) | 1 sample | Each additional sample, up to 8 per run | 10x encapsulation and library construction (per sample) |
| $620 | $50 | ||
| 10x Reagent Fee** (per sample) | $1,550 | ||
| Bioanalyzer Quality Check | $20) | ||
| Sequencing | 50% of a NextSeq High output flowcell (75 or 150 cycles): see above | ||
| * please inquire for non-BNORC or non-academic pricing | |||
| **The actual reagent fees depends on how many libraries are prepped together at the same time, but these provide a rough estimate | |||
| BNORC | Non-BNORC | Non-Academic | |
| Miscellaneous | per sample | per sample | per sample |
| BioAnalyzer Analysis (standalone) | $10 | $13 | $20 |
| Qubit quanitification (Initial set up fee) | $10 | $13.50 | $20 |
| + per sample | $1 | $1.35 | $2 |
| Drop-seq Library construction: per ~750 ul run | $600 | $750 | $1,200 |
| Tissue prep for 10x sc-RNA-seq | |||
| 10x sc-RNA-seq | |||
| Drop-seq Droplet generation: per 2-2.5ml | $600 | $800 | $1,200 |
| BNORC | Non-BNORC | ||
| Bioinformatic Consultation (per hour) | $100 | $135 | $200 |
| Per flowcell (Estimates) | |||
| Standard ChIPseq/ATACseq output (QC/alignment/browser track generation) | $300 | $400 | $600 |
| + Peak calling, library normalization, counts | $500 | $665 | $1,000 |
| + Differential enrichment/GSEA | $800 | $1,064 | $1,600 |
| + Motif Enrichment | $100 | $133 | $200 |
| Standard RNAseq output (QC/alignment/browser track generation) | $300 | $399 | $600 |
| + Transcript determination, normalization, counts | $500 | $665 | $1,000 |
| + Differential expression/GSEA | $800 | $1,064 | $1,600 |
| Single Cell RNA-seq analysis | |||
| Cell Ranger standard output (per sample) | $800 | $1,064 | $1,600 |
| Custom pipeline output with guided clustering, marker identification, figure generation (feature, dot, violin, & cluster proportion plots)* | $1200 + | please inquire | please inquire |
| * Estimate will vary depending on # cells/nuclei being analyzed | |||
| Custom analysis (per hour of bioinformatician time) | $125 | $165 | $250 |
| Longterm Storage/TB/month | $20 | $27 | $40 |


Last Updated on June 21, 2022
